To What Extent Does Life Simply Invent Itself As It Goes Along?
The evidence may surprise usAccording to a popular anti-creationist website Talk Origins, one of the strongest pieces of evidence for evolution and common descent is the phylogenetic signal. In their view, when we look at specific properties across multiple species — number of legs, presence or absence of wings, the presence of a gene, a configuration of nucleotides in the genome, etc. — such properties will form a strongly nested hierarchy. We should expect to see such a strongly nested hierarchy if organisms evolved from one another and therefore a “family tree” links all the organisms together. However, if a property that appears only on a branch, it will be shared only by that branch’s leaves. Leaves on a different branch will not have that property.
But what if a few leaves on completely separate branches share a property but the rest of the leaves on both branches do not? Such a finding is contrary to what we’d expect from the “family tree” (strongly nested hierarchy) model of evolution. However, it is quite common and it has a name: convergent evolution. For example, birds are said to have evolved ultraviolet vision at least eight times. Dolphins and insects share components of a hearing system.
Life forms cannot tell us their history. We must try to figure it out. To measure whether a collection of organisms is really a family, with a family tree, a variety of metrics has been proposed. If the collection achieves a high score on these metrics, then a family tree is inferred. The high score is called a phylogenetic signal.
But how can we be sure that the high score could not have occurred by some means other than inheritance via a family tree, for example, via convergent evolution.
Consider the letters that stand in for the nucleotides in our genomes, G, A, T, and C. If we just selected the letters randomly, some would line up in an apparently meaningful way by chance. To guard against chance similarities, various researchers have evaluated the “phylogenetic signal” metrics with random datasets, to determine how much similarity could just be chance. If the scores fall above this randomness threshold, then the phylogenetic signal is strong. Then researchers feel that they can be confident inferring a family tree.
One of the most popular metrics is the “consistency index” (CI). A 1991 paper by Klassen et. al. calculated a randomness threshold for CI, and then evaluated 70+ published CI values to determine if they fell above or below the threshold. All but 2 fell well above the threshold, showing the majority of the CI values were statistically significant and not due to random happenstance.
The authors of the paper were cautious about the conclusions that could be drawn from this result but other sources (such as the Talk Origins website) have taken their result as strong evidence for an evolutionary family tree.
Not so fast. There is a flip side to the CI metric. If we understand this flip side, we also understand why Klassen et al were a bit more circumspect in their conclusions than their fans were. The CI metric ranges from 0 to 1, with 1 being scored by a perfect tree structure. One might then expect that if organisms are the result of an evolutionary tree, they should score 1. Well, let us look at the chart from the paper:
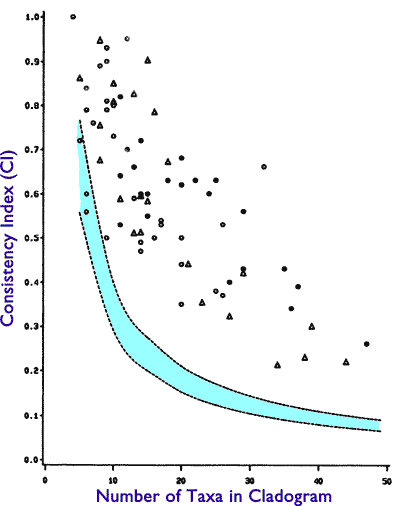
Note that only one study achieves the perfect CI score of 1, and most are well below. Yet, these values are at the same time above the random threshold. What does this mean?
That’s the flip side of the CI metric. When we subtract the CI metric from 1, we get another metric called the homoplasy index (HI). As we might expect, HI measures the opposite of CI; i.e. how untree-like the generating structure is. What does it mean to be “untree-like”? It means that properties will take big jumps across the tree, landing like a squirrel in totally unassociated branches and leaves.
If we were to diagram what that looks like (leaving out the squirrel and using only plant materials), it would look like a tree with vines connecting the branches— and sometimes with entirely new trees popping up in midair and somehow getting grafted into the main tree. It’s very unlike the traditional evolutionary tree we are taught. Instead, it is a bizarre, transdimensional, vine-twisted tree.
Let us return to the Klassen 1991 graph and imagine it flipped on its head:
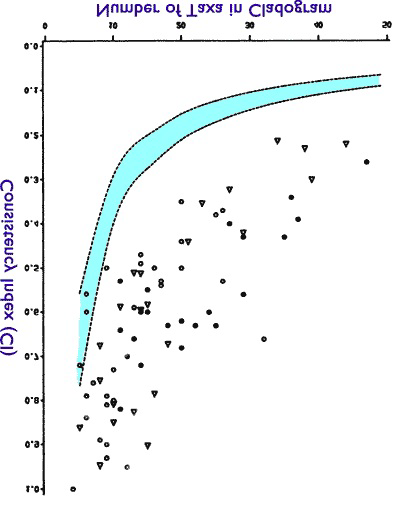
This flipped graph is the HI graph for those 70+ studies. What is it showing us? It is showing us that instead of a “phylogenetic signal,” as the scientists have studied ever larger collections of organisms, the result looks more like the bizarro tree than the evolutionary tree.
This bizarro tree is very mysterious. However, there is one other place where we see this type of bizarro tree. That place is the realm of human inventions. If we were to graph the progression from horse-drawn carriage to motorcar, we would see something that looks a bit like genealogical progress, for example, the progress in horse harness. However, we would also see the bizarre jumps where an inventor has looked entirely outside the world of horse-drawn carriages and taken inspiration from steam locomotives and steamboats. We would also see entirely separate branches of research into electronics that find their way into the internals of the modern sedan.
Let us take a step back from our presuppositions, and compare the flipped HI graph to our two possible candidate models. On the one hand we have the evolutionary tree. On the other hand we have the bizarro tree of human invention. Which model does the HI graph best indicate? It seems to be the bizarro tree, not the evolutionary tree. Does this mean that biological history looks more like a graph of invention than a graph of evolution? If so, then the “phylogenetic signal” is more appropriately named the “design signal.”
Note: Convergent evolution (unrelated life forms using the same intricate patterns) is quite common.
You may also enjoy: Evolution and artificial intelligence face the same basic problem. Think of the word ladder game, where we transform one word into another by changing only one letter at a time.
